Publications
BxChip© Clinical Tissue Array Increases Cancer Detection Rate & Amount of Tissue Available for Pathologist Review
Kirk Wojno*, Royal Oak, MI; Rima Al-Jundi, St. Clair Shores, MI; Ann Mazurco, Royal Oak, MI; Hayder Al Hamzawy, Salt Lake, UT
INTRODUCTION AND OBJECTIVES: Due to significant cuts in reimbursement, more efficient histology techniques are needed to maintain the economic viability of the histology laboratory. There is also an increasing need to maximize tissue preservation for ancillary testing. The BxChip© (Leavitt Medical Inc. Patent Pending) is a clinical tissue array made of an artificial tissue-like material which easily receives and holds multiple tissue cores. Encasing numerous needle cores, enables the arrayed samples to be processed, embedded and sectioned as though they were a single standard tissue. Colored dividers between each core allow the pathologist to distinguish different anatomic sites, while providing orientation of each core. The purpose of this study is to compare the overall prostate cancer detection rates and the average amount of tissue present (average area and core-length) on normal slides versus those made with BxChip© clinical tissue arrays.
Originally Published in The Journal of Urology®
Wojno K, Al-Jundi R, Mazurco A, Hamzawy HA. Mp 16-19 BxChip© Clinical tissue array increases cancer detection rate & amount of tissue available for pathologist review. The Journal of Urology. 2016; 195(4). doi:10.1016/j.juro.2016.02.2584
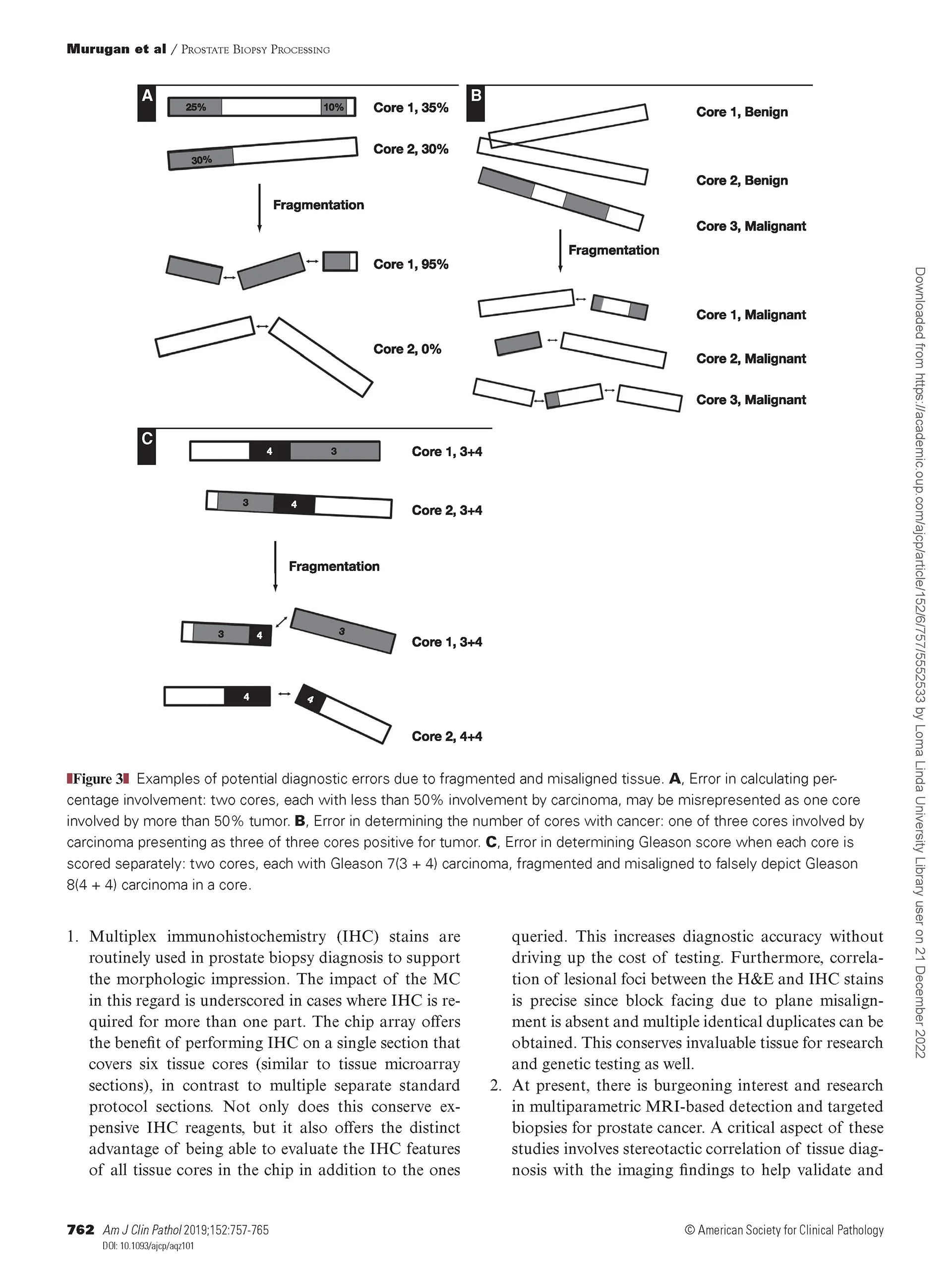
Prostate Biopsy Processing: An Innovative Model for Reducing Cost, Decreasing Test Time, and Improving Diagnostic Material
Murugan P, Shukla D, Morocho J, Smith D, Sciacca D, Pickard M, Wahlsten M, Gunderson A, Konety B,Khalifa MA, Warlick C.
OBJECTIVES, RESULTS, AND CONCLUSION:
Current protocols for processing multiple prostate biopsy cores per case are uneconomical and cumbersome. Tissue fragmentation and loss compromise cancer diagnosis. We sought to study an alternate method to improve processing and diagnosis of prostate cancer.
A significant reduction (more than threefold) in preanalytical and analytical time was observed using the multiplex method. Nonlinear fragmentation was absent, in contrast to standard processing.
The BxChip© reduced tissue fragmentation and increased efficiency of prostate biopsy diagnosis. It also resulted in overall cost savings and significantly increased tissue length.
Originally Published in The American Journal of Clinical Pathology© Murugan P, Shukla D, Morocho J, Smith D, Sciacca D, Pickard M, Wahlsten M, Gunderson A, Konety B,Khalifa MA, Warlick C. Prostate Biopsy Processing: An Innovative Model for Reducing Cost, Decreasing Test Time, and Improving Diagnostic Material, American Journal of Clinical Pathology, Volume 152, Issue 6, December 2019, Pages 757-756, https://doi.org/10.1093/ajcp/aqz101
Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer
Misop Han, Alan W. Partin, Marianna Zahurak, Steven Piantadosi, Jonathan I. Epstein and Patrick C. Walsh
Results:
With a median followup of 5.9 years (range 1 to 17) 360 men (17%) had biochemical recurrence. Overall actuarial 5, 10 and 15-year biochemical recurrence-free survival rates were 84%, 72% and 61%, respectively. The relative risk of biochemical recurrence following surgery decreased with time, even after adjusted for other perioperative parameters. Variables identified for the preoperative model were biopsy Gleason score, clinical TNM stage and PSA. Variables identified for the postoperative model were prostatectomy Gleason score, PSA and pathological organ confinement status. Nomograms were generated and corrected for the decreasing relative risk of biochemical recurrence over time.
Originally Published in the Journal of Urology
Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003 Feb;169(2):517-23. doi: 10.1097/01.ju.0000045749.90353.c7. PMID: 12544300
Prediction of Pathologic Stage Based on Clinical Stage, Serum PSA, and Biopsy Gleason Score: Partin Tables in the Contemporary Era
Jeffrey J. Tosoian, Meera Chappidi, Zhaoyong Feng, Elizabeth B. Humphreys, Misop Han, Christian P. Pavlovich, Jonathan I. Epstein, Alan W. Partin, Bruce J. Trock
The James Buchanan Brady Urological Institute and Department of Urology at the Johns Hopkins University School of Medicine, Baltimore, MD, USA
Results:
Originally Published in the Journal of BJUI International
Tosoian JJ, Chappidi M, Feng Z, Humphreys EB, Han M, Pavlovich CP, Epstein JI, Partin AW, Trock BJ. Prediction of pathological stage based on clinical stage, serum prostate-specific antigen, and biopsy Gleason score: Partin Tables in the contemporary era. BJU Int. 2017 May;119(5):676-683. doi: 10.1111/bju.13573. Epub 2016 Jul 29. PMID: 27367645.